[5th Mar 2024] Committed to MIT's EECS PhD program and will be joining in Fall 2024.
[19th Dec 2023] Preprint summarizing findings from Microsoft Research internship is under review at npj Science of Food.
[21st Aug 2023] Started my Master's in Electrical and Computer Engineering at Cornell Tech supported by a merit-based scholarship.
[5th Jun 2023] Began research internship with Aqemia, a French startup leveraging machine learning and quantum physics to accelerate the drug discovery process.
[27th May 2023] Graduated from Brown with degree in Physics and Applied Mathematics. My thesis recieved departmental honors and can be viewed here.
[1st Nov 2022] New preprint summarising findings from Microsoft Research internship available on arxiv.
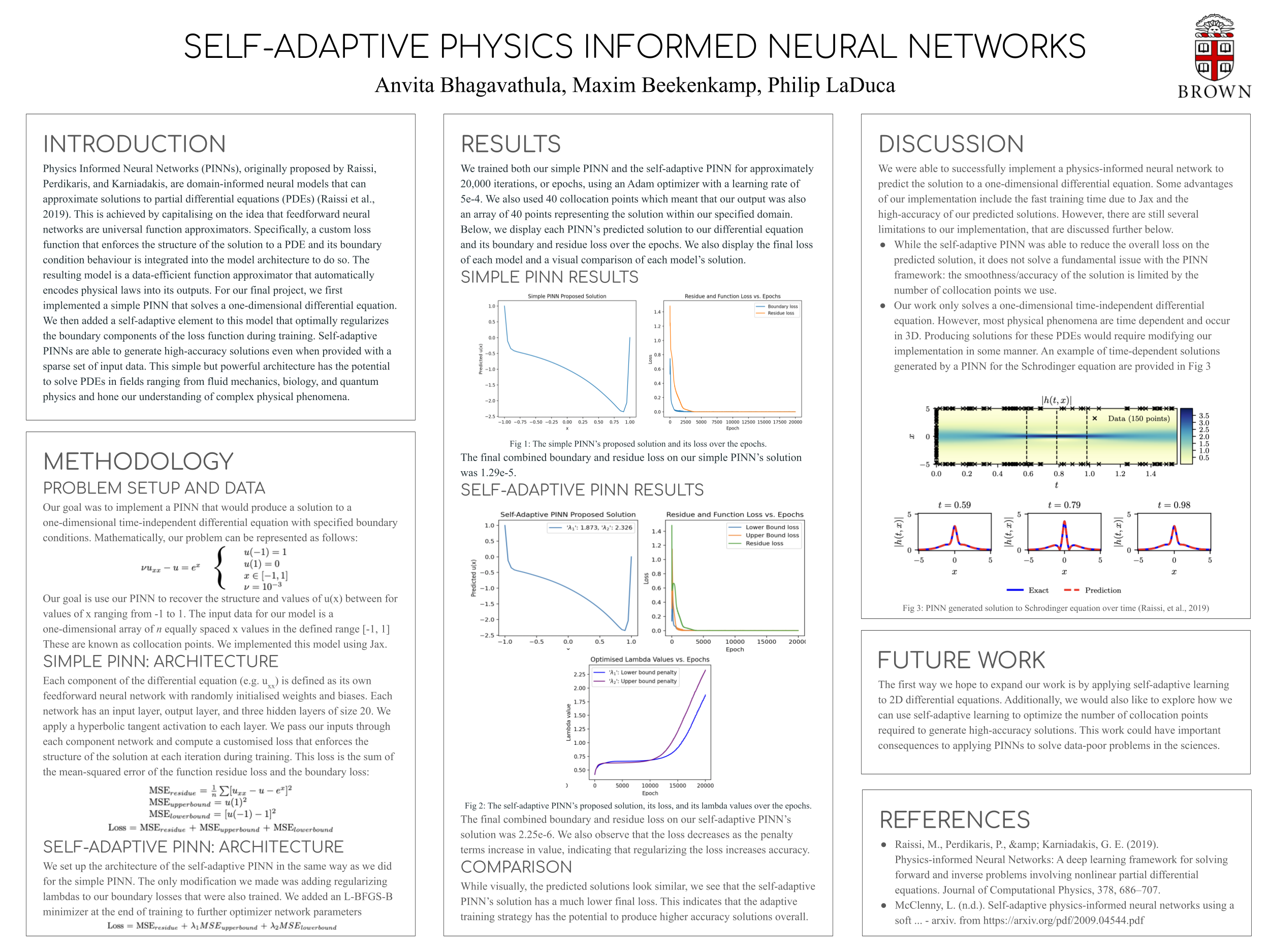
[31st Oct 2022] Started new research project with The Crunch Group at Brown researching self-adaptive physics-informed neural networks.
[26th Aug 2022] Finished my internship with Microsoft Research and filed a provisional patent for our proposed methodology to predict protein digetibility.
[6th June 2022] Started my internship with Microsoft Research where I am working with Dr. Sara Malvar and Dr. Ranveer Chandra with the Research for Industry Group.